Cellartis iPSC rCas9 Electroporation and Single-Cell Cloning System User Manual
(030619)
takarabio.com
Takara Bio USA, Inc.
Page 15 of 24
VII. Editing hiPS Cells by Electroporation
Once your sgRNA has been prepared, the electroporation protocol can begin. We recommend using the Neon
Transfection System or the Amaxa 4D-Nucleofector System to electroporate cells. Our electroporation protocol
has been optimized using the Neon Transfection System to electroporate cells from Cellartis Human iPS Cell Line
18 (Cat. No. Y00305) and Cellartis Human iPS Cell Line 22 (Cat. No. Y00325) cultured in the DEF-CS culture
system (Cat. No. Y30010). Please refer to the Neon Transfection System User Manual and manufacturer’s
website for detailed operating instructions for the Neon Transfection System.
Likewise, please refer to the Amaxa 4D-Nucleofector System manufacturer’s website for detailed operating
instructions:
http://www.lonza.com/products-services/bio-research/transfection/genome-editing.aspx
IMPORTANT:
•
Use the Cellartis DEF-CS 500 Culture System (Cat. No. Y30010) during the gene editing process as well as
the cell recovery step (prior to single-cell cloning, Section VIII).
•
Please note that when the following protocol specifies use of “COAT-1,” it is referring to the DEF-CS
COAT-1 from the Cellartis DEF-CS 500 Culture System. Do not use the coating contained in the Cellartis
iPSC rCas9 Electroporation and Single-Cell Cloning System, which is abbreviated as “SCC-COAT-1” due to
its sole use for the single-cell cloning and expansion protocols.
•
If cells have not been previously adapted to growing in the DEF-CS system, we strongly recommend
transitioning cells by passaging five times in the DEF-CS system prior to performing the editing experiments.
A.
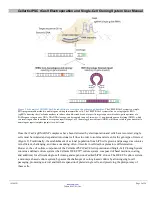
Protocol Overview
Figure 8. Workflow for electroporation-based delivery of rCas9/sgRNA complexes.
B.
Preparing Media and Cells
Prior to beginning the electroporation protocol, prepare a starting culture of hiPSCs. You will need
1.5 x 10
5
cells per sample to be electroporated. (Please see the NOTE under Section VII.B.3 for details.)
Electroporating Cells
rCas9/sgRNA complexes are electroporated into target cells.
Preparing the rCas9/sgRNA Complex
rCas9 protein and sgRNA are incubated to facilitate complex formation.
Preparing Media and Cells
Medium and plates are prepared for use after electroporation.
Cells are washed and suspended in an appropriate buffer.