Cellartis iPSC rCas9 Electroporation and Single-Cell Cloning System User Manual
(030619)
takarabio.com
Takara Bio USA, Inc.
Page 5 of 24
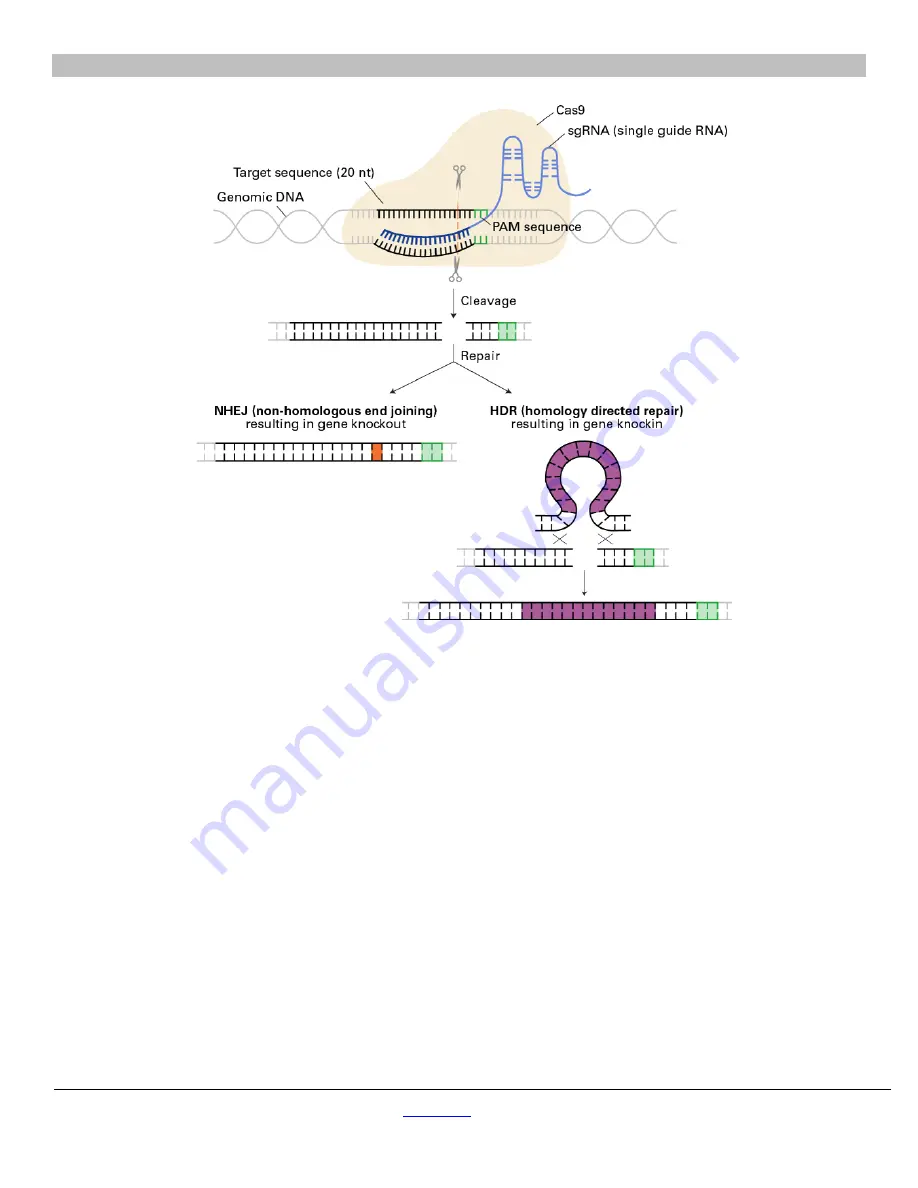
Figure 1. Schematic of CRISPR/Cas9-based editing to create gene knockouts and knockins.
The CRISPR/Cas9 system is a simple,
RNA-programmable method to mediate gene editing in mammalian cells. The CRISPR/Cas9 system relies on a single guide RNA
(sgRNA) directing the rCas9 endonuclease to induce a double-strand break at a specific target sequence three base-pairs upstream of a
PAM sequence in genomic DNA. This DNA cleavage can be repaired in one of two ways: 1) non-homologous end joining (NHEJ), which
can result in gene knockout due to error prone repair (orange), or 2) homology directed repair (HDR), which can result in gene knockin if a
homologous repair template (purple) is co-delivered.
Once the rCas9/sgRNA RNP complexes have been delivered by electroporation and cells have recovered, single
cells must be isolated and expanded into clonal cell lines in order to isolate and screen for the genotype of interest
(Figure 2). Traditionally, the establishment of a clonal population from hiPS cells grown and passaged as colonies
is inefficient, challenging, and time-consuming; often, it results in cell death or premature differentiation.
However, the cell culture component of the Cellartis iPSC rCas9 Electroporation and Single-Cell Cloning System
contains a defined culture system (the Cellartis DEF-CS™ culture system, composed of basal medium, coating,
and additives) for efficient single-cell cloning and expansion of edited hiPSC clones. The DEF-CS culture system,
a monolayer-based culture system, bypasses the challenges of colony-based culture by allowing single-cell
passaging, promoting survival and further expansion of plated single cells, and preserving the pluripotency of
these cells.